Table of Contents
- Introduction
- Invitation to participate
- Why you should participate
- What does it take?
- Description of participating systems
- Participate
Introduction
In light of the COVID-19 pandemic, there has been a rapid increase in SARS-CoV-2-related research, accompanied by the emergence of a number of literature datasets [33166392][CORD-19], bioinformatics resources [33147627] and text mining tools/systems [33279995] developed specifically for SARS-CoV-2 and COVID-19 research.
The motivation for this track is to provide developers with a more formal evaluation about the usefulness of their tools by providing user-in-the-loop feedback.
In addition, the track aims to promote text mining web applications conceived to support COVID-19 research. Similar to previous interactive tasks (e.g., 27589961), these TM systems will be reviewed by the appropriate users, providing feedback on effectiveness and usability.
Invitation to Participate
This is an open invitation to participate in exploring one or more COVID 19 related text mining systems of your choice any time during the period September 1 to September 30 prior to the BioCreative VII workshop (November 8-10, 2021). The activity is conducted remotely and is time flexible. It includes two subtasks: one guided, which involves following pre-defined tasks, and another exploratory, which involves using your own examples and trying functionalities of interest. You will provide feedback about the system via a survey.
The collective feedback gathered will be sent to the developers and the results will be presented during the BioCreative VII virtual workshop on November 8-10, 2021. Attendance to the workshop is not a prerequisite for participation in this activity, but you are more than welcome to do so (it is free).
Why you should participate
The benefits to participants of this activity are multifold, including:
- direct communication and interaction with developers
- exposure to new text mining tools that can be potentially adapted and integrated into your workflow
- contribution to the development of text mining systems that meet the needs of the community
What does it take ?
The time frame is very flexible you can try one or more systems of your interest any time between September 1 and September 30. We expect that reviewing a system may take 1-2 hours and completing the survey 30 min. Your participation involves selecting a system to review and provide feedback by completing two activities and surveys found in the link under "Choose system to review".
1-Guided: You will go to the review page for the system, some tutorial/instruction on the system's capabilities. You will also be given an example to walk through. You will report on the finding via a survey at the end of the guided and exploratory activity.
2-Exploratory: Navigate the system and try your own examples, you will report on findings via a survey at the end of the activity.
Description of Participating Systems
Below is the list of the systems that are participating in the upcoming BioCreative VII interactive demo track (Track 4). Also note that the information in the table is limited to that pertaining to the task proposed for the interactive activity.
To participate in reviewing one or more of these systems, just click on link under "Choose system to review" in table below.
| System | Description | Short Overview | Choose system to review |
|---|---|---|---|
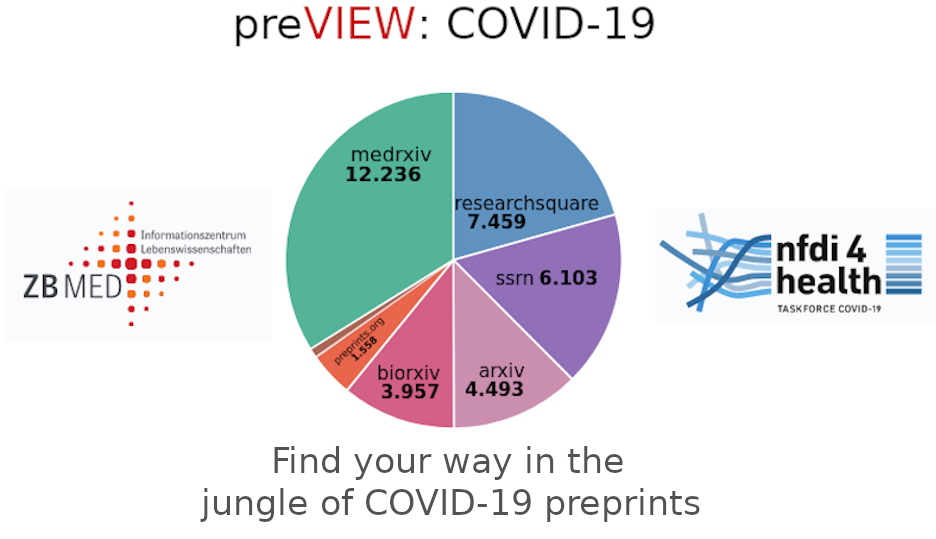 | The increased importance of preprints for COVID-19 research initiated the design of the preprint search engine preVIEW. It is a lightweight semantic search engine focusing on easy inclusion of specialized COVID-19 textual collections and provides a user friendly web interface for semantic information retrieval. | ||
AGATHA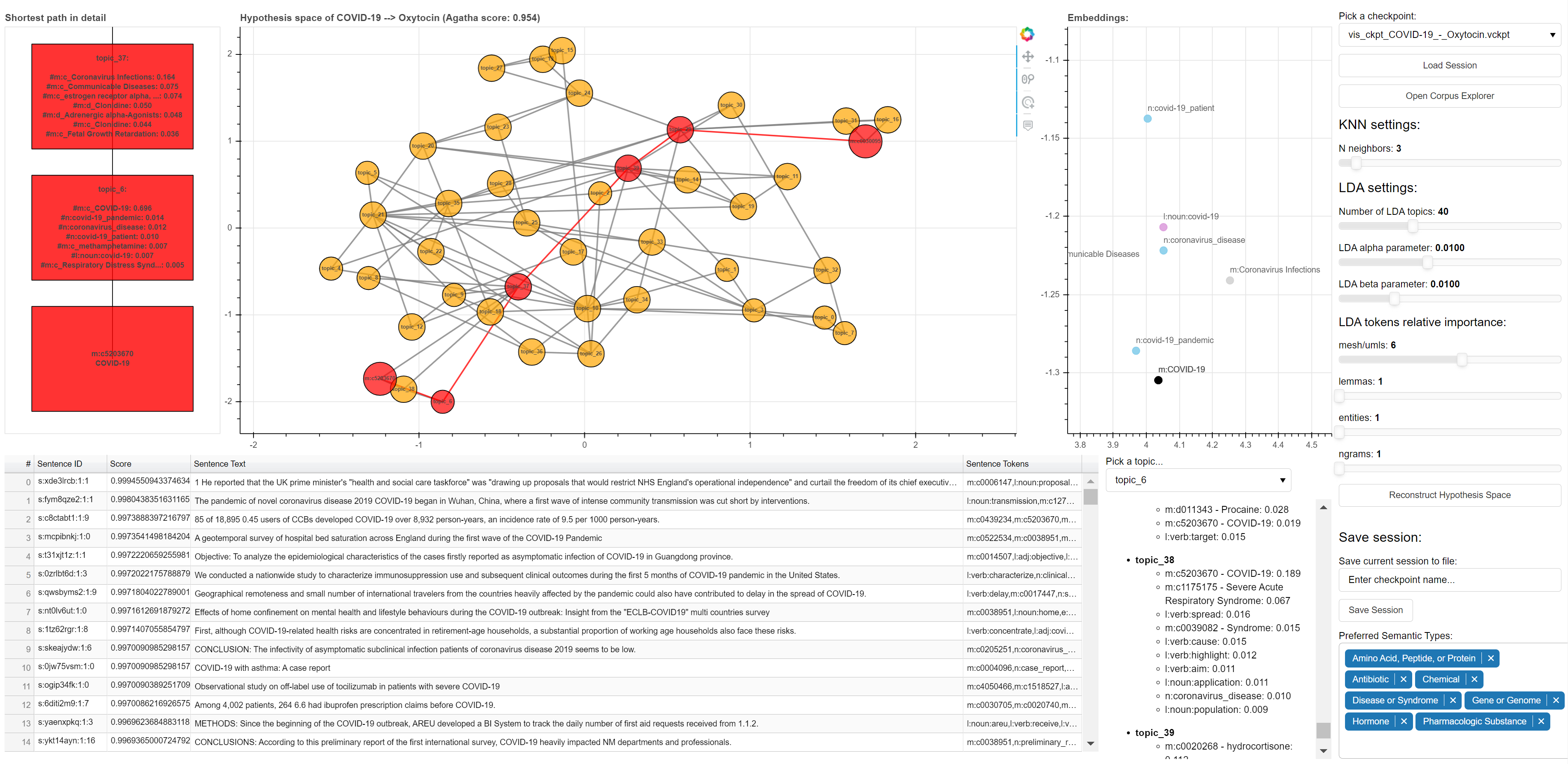 | Designed for finding implicit, undiscovered connections between avariety of biomedical concepts. Automated general purpose hypothesis generation system for COVID-19 research based on graph-mining and the transformer model. | ||
 | Biomedical Knowledge Discovery Engine. A platform for biomedical researchers to retrieve, visualize, manage and mine knowledge in scientific literature. Currently BioKDE has two main modules: search engine and knowledge graph-based visualization. | ||
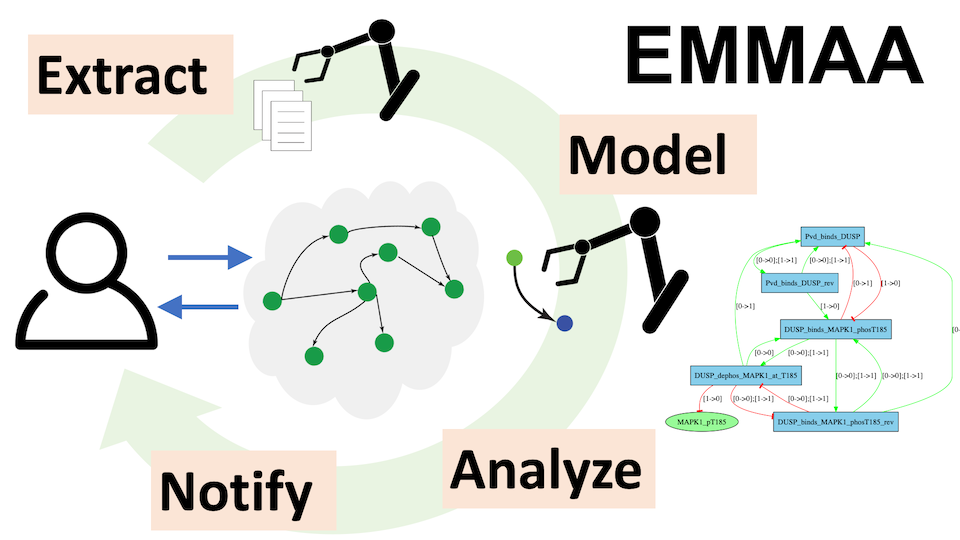 | Ecosystem of Machine-maintained Models with Automated Analysis. A framework for keeping a set of disease-related models up to date using the latest results from the scientific literature. It has been applied to modeling COVID-19 mechanisms. | ||
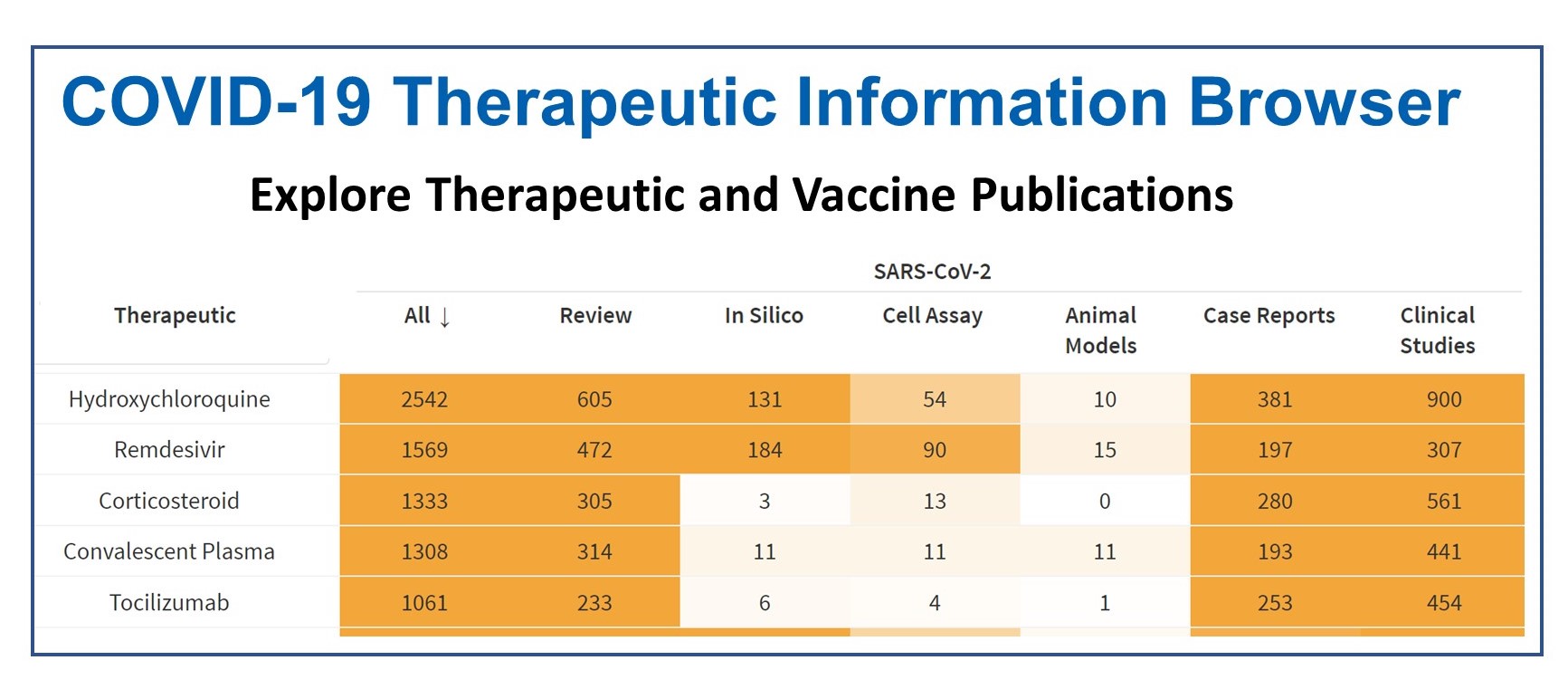 | Dashboard to help the biomedical community easily find and track scientific research about potential COVID-19 therapeutics and vaccines. |  | |
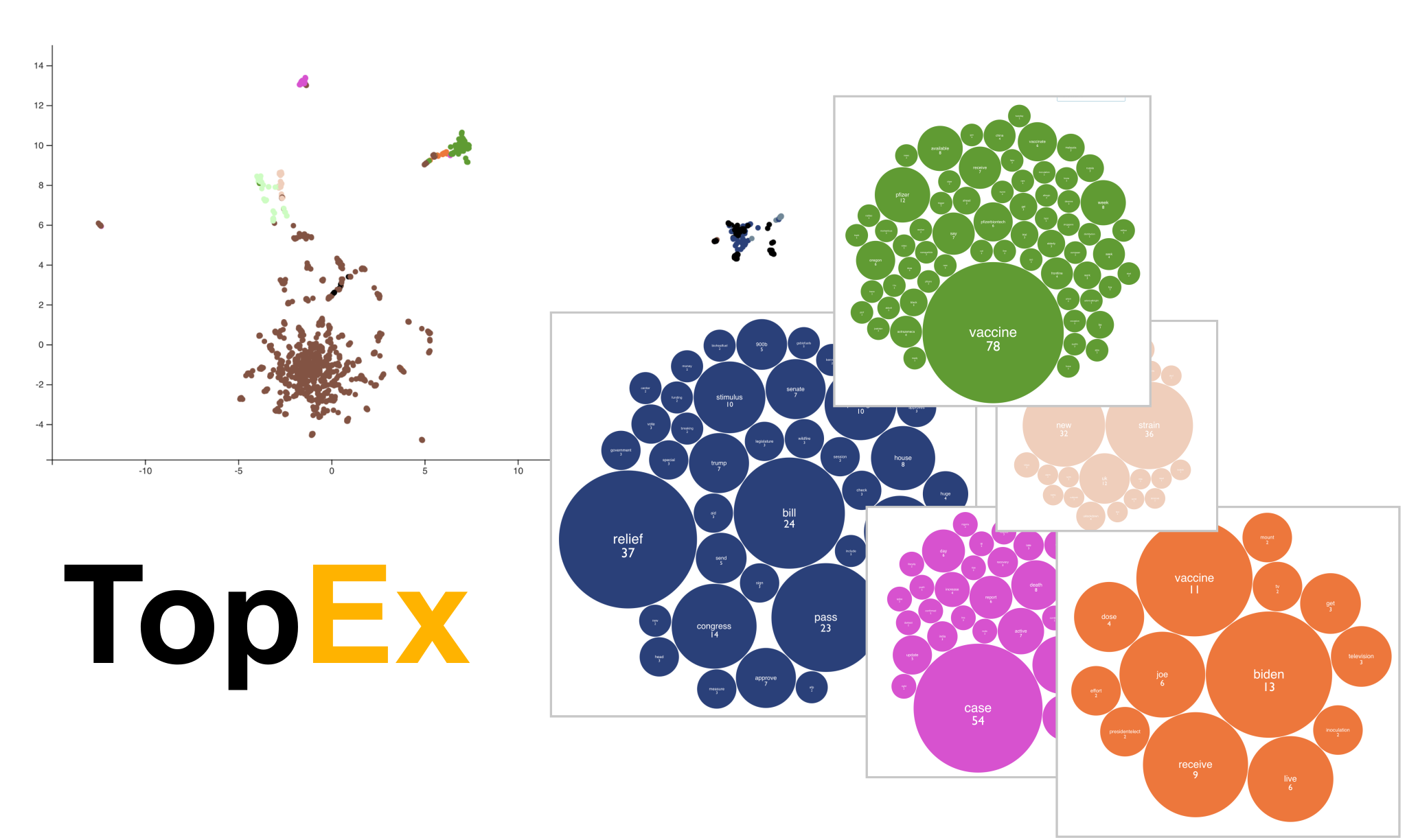 | A topic discovery and clustering tool built to aid in the exploration of topics present in a set of texts through a user interface that requires no programming or NLP knowledge. | ||
COVID19 SCAIView | Search engine for semantic searches in large text collections, that combines free text searches with the ontological representations of entities. |
You can check description of Track 4 activity for systems here
Participate
To participate, go to the "description of participating systems" table (above), and simply click on the "magnifier glass" icon for the system of interest under column "choose system to review" . This will take you to the page for the interactive activity for such system. Remember that we need your feedback by September 30.
You can always contact us if you have additional questions by sending email to Biocreative with SUBJECT TRACK4.





